Integration with bhmbasket
Kevin Kunzmann
Source:vignettes/bhmbasket-integration.Rmd
bhmbasket-integration.Rmd
library(oncomsm)
library(dplyr, warn.conflicts = FALSE)
library(future) # parallel processing
m_cores <- if (as.logical(Sys.getenv("NOT_CRAN", unset = "FALSE"))) {
parallel::detectCores()
} else {
2L # need to restrict parallel processing on CRAN
}
plan(multisession) # instruct future how to run in paralleltl;dr: The bhmbasket
package implements Bayesian hierarchical methods for basket trials with
binary endpoints. Although oncomsm does not support
hierarchical modelling, bhmbasket can be used to define
‘go’ criteria based on hierarchical models.
Start by defining a prior specification for a multi-group trial.
mdl <- create_srpmodel(
A = define_srp_prior(
p_mean = 0.3, p_n = 20,
median_t_q05 = c(3, 2, 6) - 1,
median_t_q95 = c(3, 2, 6) + 1,
recruitment_rate = 2
),
B = define_srp_prior(
p_mean = 0.4, p_n = 20,
median_t_q05 = c(2.5, 8, 18) - 1,
median_t_q95 = c(2.5, 8, 18) + 1,
recruitment_rate = 2
),
C = define_srp_prior(
p_mean = 0.5, p_n = 20,
median_t_q05 = c(2, 12, 24) - 1,
median_t_q95 = c(2, 12, 24) + 1,
recruitment_rate = 2
)
)Defining ‘go’ decision
The bhmbasket package implements dynamic borrowing
between arms in basket trials via Bayesian hierarchical models (BHMs).
Here, we demonstrate how oncomsm and the prior specified in
mdl can be used to derive probability of ‘go’ based on a
bhmbasket analysis. We use the “Berry” type model for
analysis of the response data and use a posterior \(0.25\) quantile of the response probability
above \(0.3\) to declare ‘go’. This
means that an individual arm is further developed if there is a \(0.75\) posterior probability according to
the BHM that the response rate is larger than \(0.3\).
go <- function(model, data, nsim = 250) {
set.seed(2340239L)
# convert data to bhmbasket format (multi-state to binary counts)
data <- data %>%
group_by(group_id, subject_id) %>%
summarize(
responder = any(state == "response"),
.groups = "drop_last"
) %>%
summarize(
r = sum(responder),
n = n(),
) %>%
{
bhmbasket::createTrial(.$n, .$r)
}
# define Berry model in bhmbasket
prms_berry <- bhmbasket::setPriorParametersBerry(
mu_mean = bhmbasket::logit(0.25),
mu_sd = 1,
tau_scale = 1
)
# perform analysis in bhmbasket
res <- suppressMessages(bhmbasket::performAnalyses(
scenario_list = data,
method_names = "berry",
evidence_levels = 1 - 0.25,
prior_parameters_list = prms_berry,
target_rates = c(.2, .3, .3),
n_mcmc_iterations = nsim,
verbose = FALSE
))
# 'go' if posterior quantile of response rate is sufficiently large
return(tibble(
group_id = model$group_id,
go = res$scenario_1$quantiles_list$berry[[1]][5, 2:4] >= .3
))
}Probability of ‘go’ before start of the trial
The ‘go’ criterion can then be applied to each of the resampled data sets.
tbl_decisions <- simulate_decision_rule(mdl,
c(40, 40, 40),
go,
nsim = 250,
seed = 32487)
tbl_pr_go_planning <- tbl_decisions %>%
group_by(group_id) %>%
summarize(`Pr[go] planning` = mean(go))
tbl_pr_go_planning
#> # A tibble: 3 × 2
#> group_id `Pr[go] planning`
#> <chr> <dbl>
#> 1 A 0.252
#> 2 B 0.648
#> 3 C 0.9Update probability of ‘go’
Now assume that some interim data is available. The data is fairly extreme for both group A and group B.
tbl_interim <- tribble(
~subject_id, ~group_id, ~t, ~state,
"s1", "A", 0, "stable",
"s1", "A", 1.5, "stable",
"s1", "A", 2.25, "response",
"s2", "A", 1, "stable",
"s2", "A", 2, "response",
"s3", "A", 3, "stable",
"s3", "A", 4.5, "response",
"s4", "B", 0, "stable",
"s5", "B", 1.5, "stable",
"s6", "C", 0, "stable",
"s6", "C", 1.5, "progression",
"s7", "C", 2.25, "stable",
"s7", "C", 3, "progression",
"s8", "C", 2, "stable",
"s8", "C", 3, "progression",
"s9", "C", 2, "stable",
"s9", "C", 2.6, "stable",
"s9", "C", 3, "progression",
"s10", "C", 3, "stable",
"s10", "C", 5, "progression"
)
# plot it
visits_to_mstate(tbl_interim, mdl) %>%
plot_mstate(mdl, relative_to_sot = FALSE)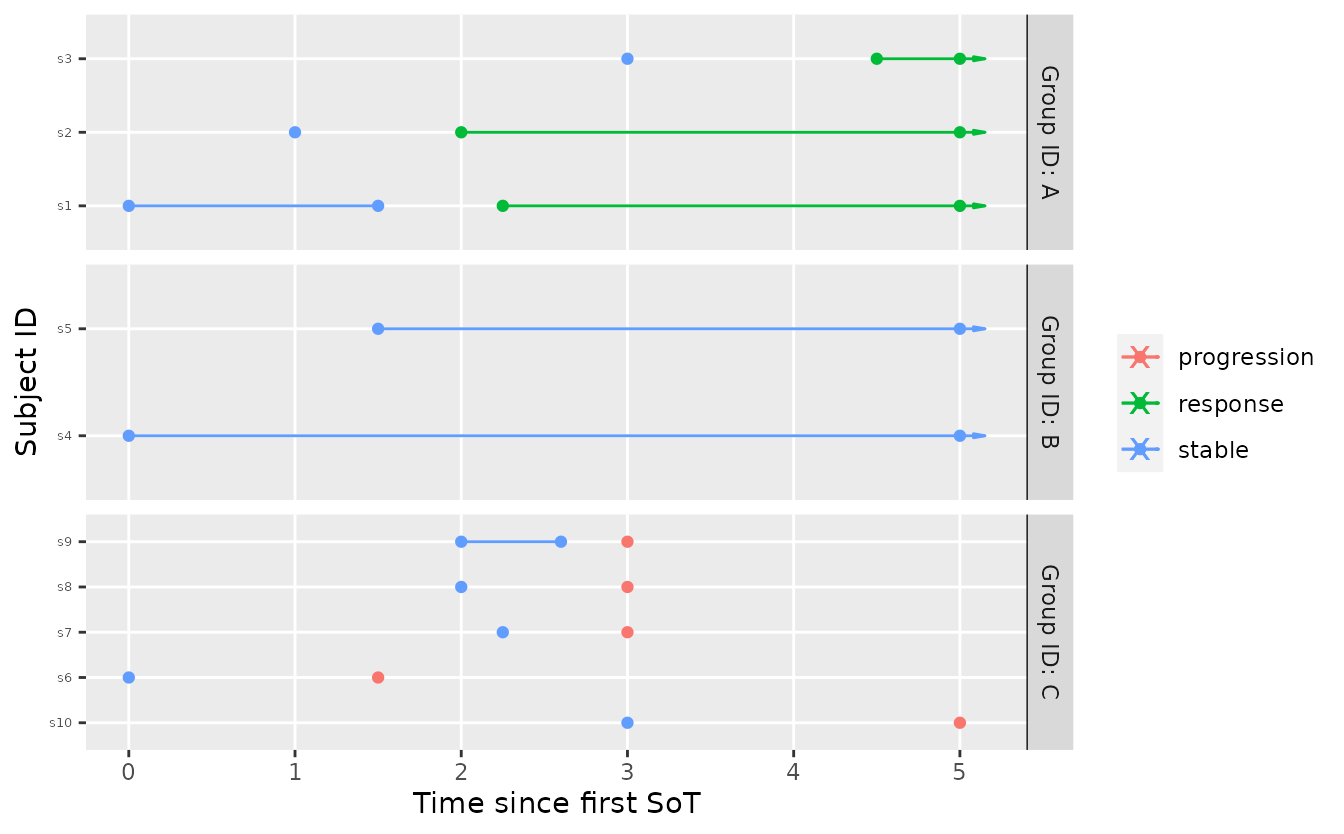
The prior can now be updated with this data.
smpl_prior <- sample_prior(mdl, seed = 2314513)
smpl_posterior <- sample_posterior(mdl, tbl_interim, seed = 2314)
tibble(
group_id = mdl$group_id,
prior = rstan::extract(smpl_prior, "p")[[1]] %>% colMeans(),
posterior = rstan::extract(smpl_posterior, "p")[[1]] %>% colMeans()
)
#> # A tibble: 3 × 3
#> group_id prior posterior
#> <chr> <dbl> <dbl>
#> 1 A 0.300 0.392
#> 2 B 0.400 0.383
#> 3 C 0.498 0.400The probability of ‘go’ is then updated by sampling forward from the posterior predictive.
tbl_decisions_interim <- simulate_decision_rule(
mdl,
c(40, 40, 40),
go,
data = tbl_interim,
nsim = 250,
seed = 32487
)
tbl_pr_go_planning %>%
left_join(
tbl_decisions_interim %>%
group_by(group_id) %>%
summarize(
`Pr[go] interim` = mean(go)
),
by = "group_id"
)
#> # A tibble: 3 × 3
#> group_id `Pr[go] planning` `Pr[go] interim`
#> <chr> <dbl> <dbl>
#> 1 A 0.252 0.608
#> 2 B 0.648 0.632
#> 3 C 0.9 0.528