Binary Endpoints in Bayesian MCP-Mod
2026-02-15
Source:vignettes/binary_endpoint.Rmd
binary_endpoint.RmdShow code
suppressPackageStartupMessages({
library(BayesianMCPMod)
library(RBesT)
library(DoseFinding)
library(dplyr)
})
set.seed(7015)
display_params_table <- function(named_list) {
round_numeric <- function(x, digits = 3) if (is.numeric(x)) round(x, digits) else x
tbl <- data.frame(
Name = names(named_list),
Value = I(lapply(named_list, function(v) {
if (inherits(v, "Date")) v <- as.character(v)
if (!is.null(names(v))) paste0("{", paste(names(v), v, sep="=", collapse=", "), "}")
else v
}))
)
tbl$Value <- lapply(tbl$Value, round_numeric)
knitr::kable(tbl)
}Introduction
This vignette demonstrates the application of the
BayesianMCPMod package for a binary endpoint. A more
detailed introduction is provided for the setting of a continuous
endpoint (analysis example
vignette).
Binary endpoints require modeling on the logit scale. We will use the
migraine dataset from the DoseFinding package as our
working example, which contains response rates after migraine treatment.
The prior (for the control group) will be based on historical trial
data.
This package makes use of the future framework for parallel processing, which can be set up as follows:
future::plan(future::multisession)Scale conventions used in this vignette
Scale conventions
- Internally, BayesianMCPMod fits binary endpoints on the logit scale.
-
probability_scale = TRUEcontrols whether outputs (summaries, predictions, plots) are transformed back to probabilities. -
deltafor MED is interpreted on the probability scale whenprobability_scale = TRUE.
Calculation of a MAP Prior
In a first step, a meta analytic prior will be calculated. This prior is based on trials results for (Diener et al. 2011), (Ho et al. 2008) and (Hewitt et al. 2011). Here we assume the following historical results for the control group. Please note that only information from the control group will be integrated leading to an informative mixture prior for the control group, while for the active groups a non-informative prior will be specified.
Show code
Our approach to establish a MAP prior is conducted in 3 steps. First
the information from the historical trials is used to establish a beta
mixture MAP prior (family=binomial).
In a next step this prior is robustified. Finally, since the
BayesianMCPMod procedure for binary endpoints requires a prior on the
logit scale, we translate this prior to this scale via sampling from the
distribution, transitioning the individual results to the logit scale
and approximating via fitting of normal mixtures of conjugate
distributions. Please note that there would be various other options to
establish a reasonable informative prior in this setting.
Show code
dose_levels <- c(0, 2.5, 5, 10, 20, 50, 100, 200)
#i) Establish MAP prior (beta mixture distribution)
set.seed(7015) # re-set seed only for this example; remove in your analysis script
map <- gMAP(
cbind(r, n - r) ~ 1 |
study,
family = binomial,
tau.dist = "HalfNormal",
tau.prior = 0.5,
beta.prior = (1 / sqrt(0.1 * 0.9)),
warmup = 1000,
iter = 10000,
chains = 2,
thin = 1
)
#> Assuming default prior location for beta: 0
map
#> Generalized Meta Analytic Predictive Prior Analysis
#>
#> Call: gMAP(formula = cbind(r, n - r) ~ 1 | study, family = binomial,
#> tau.dist = "HalfNormal", tau.prior = 0.5, beta.prior = (1/sqrt(0.1 *
#> 0.9)), iter = 10000, warmup = 1000, thin = 1, chains = 2)
#>
#> Exchangeability tau strata: 1
#> Prediction tau stratum : 1
#> Maximal Rhat : 1
#>
#> Between-trial heterogeneity of tau prediction stratum
#> mean sd 2.5% 50% 97.5%
#> 0.2710 0.2210 0.0106 0.2160 0.8290
#>
#> MAP Prior MCMC sample
#> mean sd 2.5% 50% 97.5%
#> 0.1190 0.0494 0.0454 0.1130 0.2340
prior <- automixfit(map) #fits mixture distribution from MCMC samples from above
#ess(prior)
p <- summary(prior)[1]
#ii) Robustify prior
prior.rob <- RBesT::robustify(priormix = prior,
mean = 0.5,
weight = 0.4)
#ess(prior.rob)
#iii) Translate prior to logit scale (to approximate via normal mixture model)
r <- rmix(prior.rob, n = 1e4)
log.r <- RBesT::logit(r)
prior.ctr <- automixfit(log.r, type = "norm")
sigma(prior.ctr) <- sqrt(1 / (p * (1 - p)))
#ess(prior.ctr, sigma = sqrt(1/(p*(1-p))))
#Specification of reference scale (this follows the idea of [@Neuenschwander2016]).
#Specify a prior list
prior_trt <- RBesT::mixnorm(
comp1 = c(
w = 1,
m = logit(summary(prior)[1]),
n = 1
),
sigma = sqrt(1 / (p * (1 - p))),
param = "mn"
)
prior_list <- c(list(prior.ctr), rep(x = list(prior_trt), times = length(dose_levels[-1])))
dose_names <- c("Ctr", paste0("DG_", seq_along(dose_levels[-1])))
names(prior_list) <- dose_namesDose-Response Model Shapes
Candidate models are specified on the parameter scale using the
{DoseFinding} package. We will create a Mods object, which
will be used in the remainder of the vignette. Please note that the
models are specified on the logit scale.
Show code
plot(models)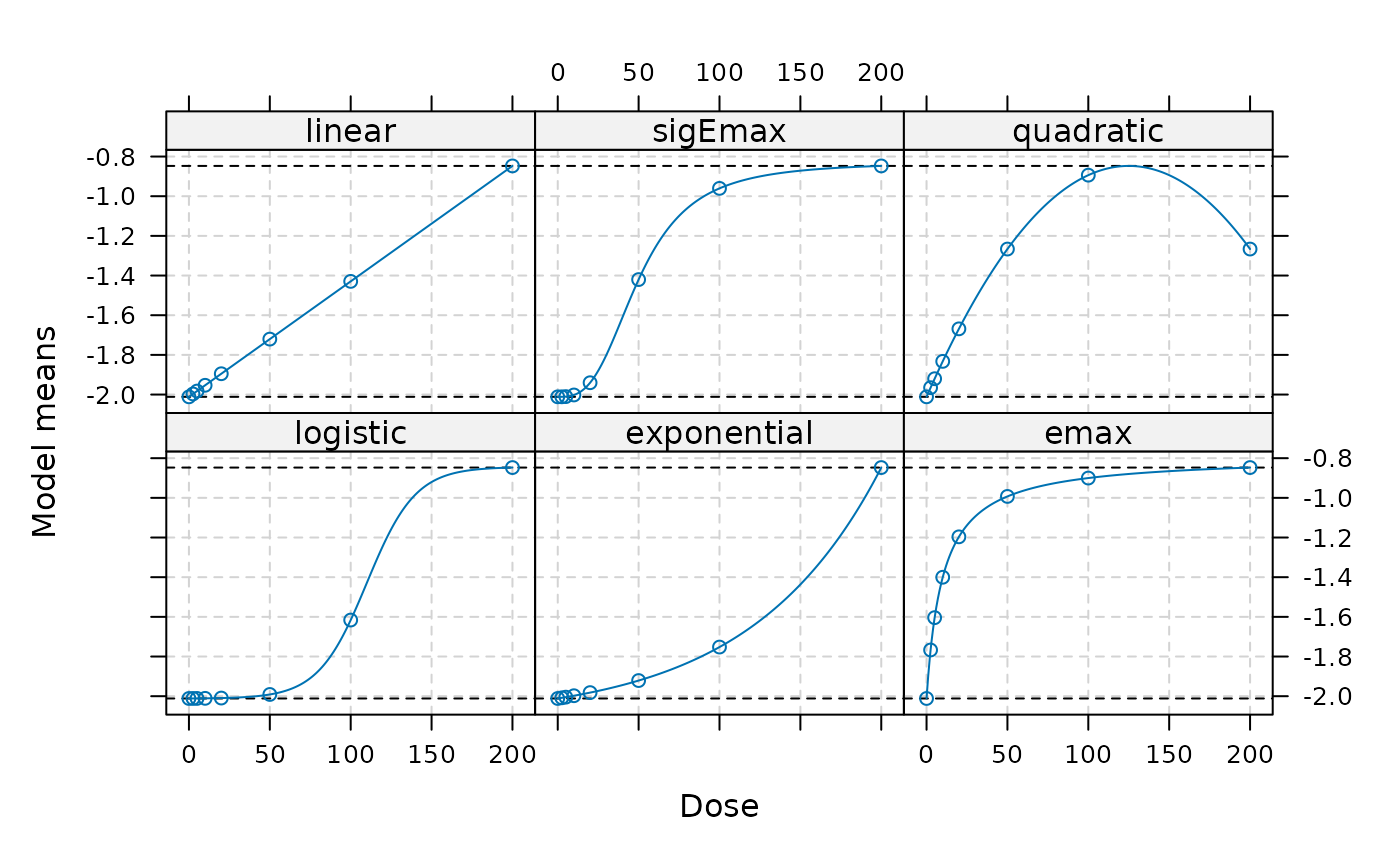
Trial Data
We will use the trial data from the migraine data set available in the DoseFinding package as our phase 2 trial data. We will apply a logistic regression (without any additional covariates) to get estimates on the logit scale.
Show code
data("migraine") #example data "migraine" from DoseFinding package
dosesFact <- as.factor(dose_levels)
N <- migraine$ntrt
RespRate <- migraine$painfree/N
##Execution of logistic regression and readout of parameters
## (please note that estimates are automatically on logit scale)
logfit <- glm(RespRate ~ dosesFact - 1, family = binomial, weights = N)
muHat <- coef(logfit)
S <- vcov(logfit)Posterior Calculation
In the first step of Bayesian MCPMod, the posterior is calculated by combining the prior information with the estimated results of the trial (Fleischer F 2022).
The summary of the posterior can be provided on the probability scale.
Show code
PostLogit<-getPosterior(prior_list,mu_hat=muHat,S_hat=S)
summary(PostLogit,probability_scale=TRUE)
#> mean sd 2.5% 50.0% 97.5%
#> Ctr 0.1071168 0.02127962 0.06930686 0.1058282 0.1520040
#> DG_1 0.1359001 0.06177459 0.04833967 0.1248126 0.2859179
#> DG_2 0.1222136 0.05057452 0.04865093 0.1137505 0.2436500
#> DG_3 0.2562256 0.05432101 0.16104931 0.2524449 0.3726678
#> DG_4 0.1943149 0.04961478 0.11121906 0.1895612 0.3041997
#> DG_5 0.2184915 0.05083942 0.13146633 0.2142515 0.3293963
#> DG_6 0.2401204 0.05496681 0.14521827 0.2358212 0.3591977
#> DG_7 0.3618200 0.06188555 0.24769337 0.3594987 0.4889697Bayesian MCPMod Test Step
The testing step of Bayesian MCPMod is executed using a critical value on the probability scale and a pseudo-optimal contrast matrix.
A contrast matrix is generated based on the number of patients per dose group (see (Fleischer F 2022) for more details). Please note that here also other options would be possible (e.g. using weight based on the observed variability).
The critical value is calculated using (re-estimated) contrasts for frequentist MCPMod to ensure error control when using weakly-informative priors.
Show code
contr_mat_prior <- getContr(
mods = models,
dose_levels = dose_levels,
dose_weights = N)
set.seed(7015) # re-sets seed only for this example; remove in your analysis script
crit_pval <- getCritProb(
mods = models,
dose_levels = dose_levels,
cov_new_trial = S,
alpha_crit_val = 0.05
)The Bayesian MCP testing step is then executed:
Show code
BMCP_result <- performBayesianMCP(
posterior_list = PostLogit,
contr = contr_mat_prior,
crit_prob_adj = crit_pval)Here as well it should be noted that this evaluation happens on the logit scale.
Summary information:
BMCP_result
#> Bayesian Multiple Comparison Procedure
#> Significant: 1
#> Critical Probability: 0.9790239
#> Maximum Posterior Probability: 0.99999
#> Posterior Probabilities for Model Shapes
#> lin sigE quad log exp emax
#> Posterior Prob 0.9999900 0.9999338 0.9998325 0.9999509 0.9999644 0.9999885
#> Significant 1 1 1 1 1 1The testing step is significant, indicating a non-flat dose-response shape. All models are significant.
Model Fitting and Visualization
In the model fitting step the posterior distribution is used as basis.
Both simplified and full fitting can be performed. Here we are focusing on the simplified fit. Furthermore we specify that the fit should be provided on the probability scale for easier interpretation of results.
The output of the fit includes information about the predicted effects for the included dose levels, the generalized AIC, and the corresponding weights.
Show code
##Perform modelling
model_fits <- getModelFits(
models = models,
dose_levels = dose_levels,
posterior = PostLogit,
simple = TRUE,
probability_scale = TRUE)Plots of fitted dose-response models and an AIC-based average model including credible bands (orange shaded areas, default levels are 50% and 95%).:
#Default is on probability scale
plot(model_fits,cr_bands = TRUE)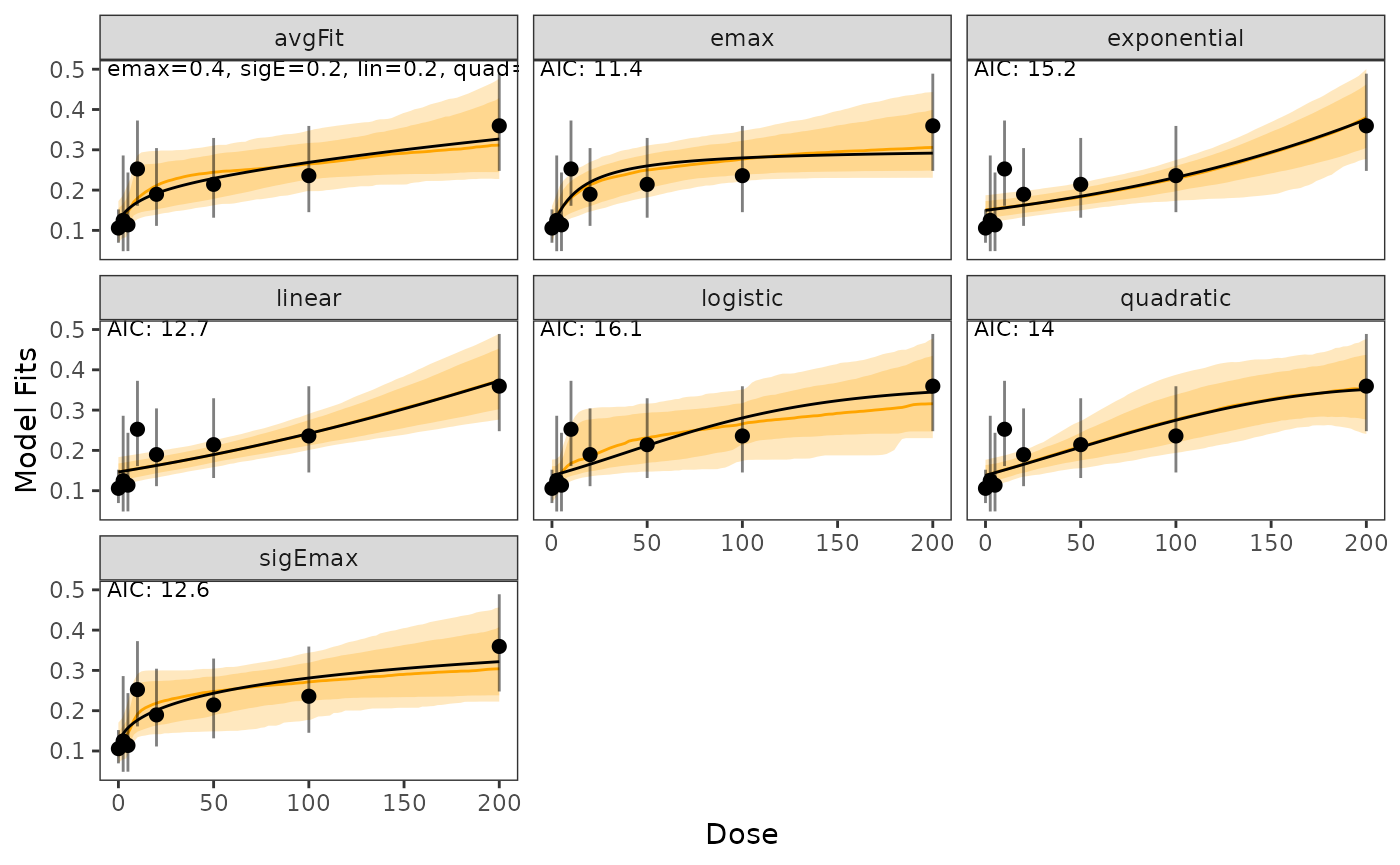
In case models should be shown on the logit scale this can be done in the following way:
plot(model_fits,probability_scale=FALSE)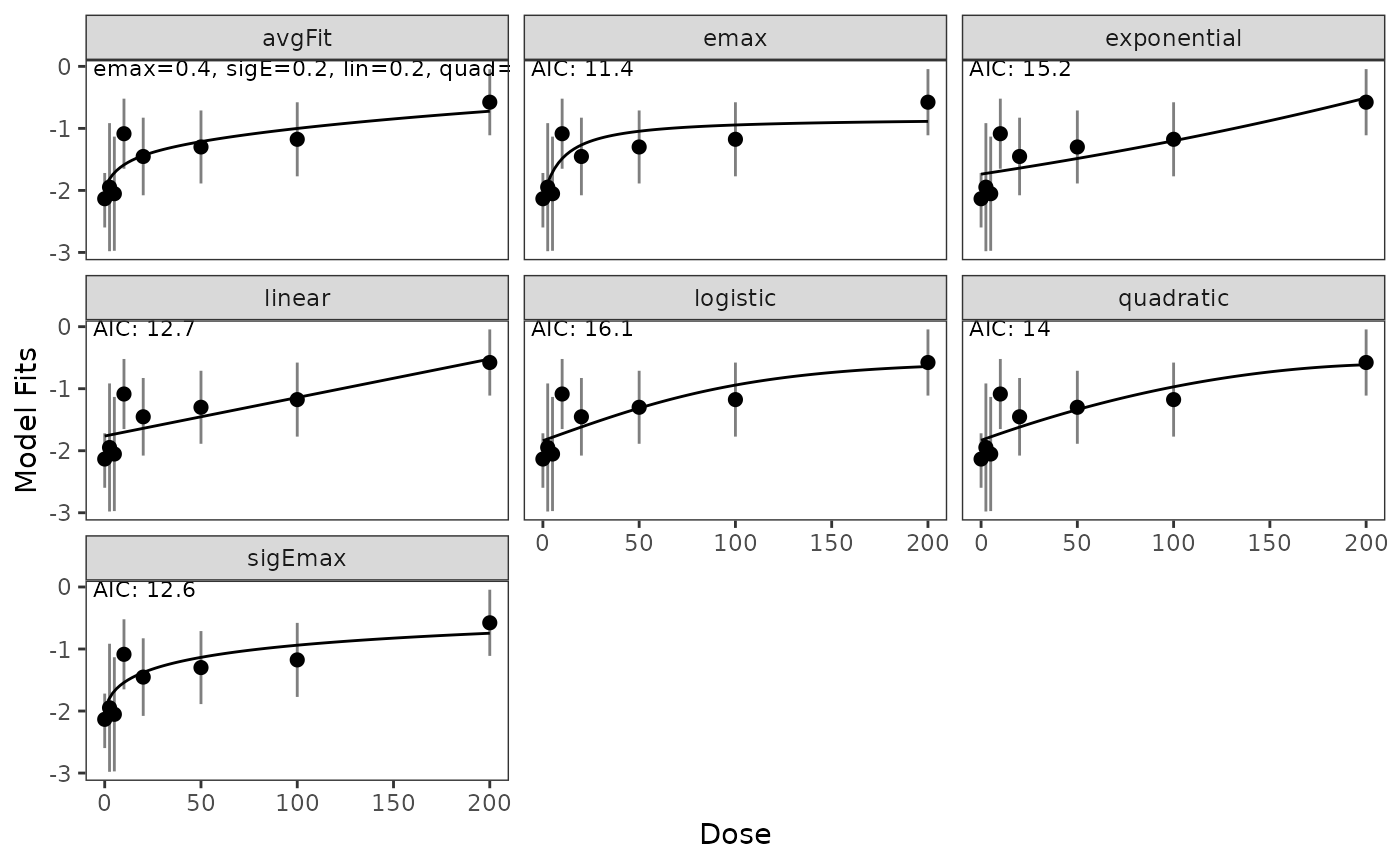
Estimates (also for dose levels not included in the trial) can be shown via:
| Name | Value | |
|---|---|---|
| avgFit | avgFit | 0.120, 0.140, 0.170, 0.300, 0.327 |
| emax | emax | 0.106, 0.133, 0.184, 0.288, 0.292 |
| exponential | exponential | 0.150, 0.151, 0.156, 0.294, 0.375 |
| linear | linear | 0.146, 0.148, 0.154, 0.303, 0.372 |
| logistic | logistic | 0.137, 0.141, 0.151, 0.323, 0.345 |
| quadratic | quadratic | 0.138, 0.141, 0.151, 0.325, 0.351 |
| sigEmax | sigEmax | 0.107, 0.143, 0.176, 0.305, 0.322 |
The bootstrap-based quantiles can also be directly calculated via the
getBootstrapQuantiles() function and a sample from the
model fits can be bootstrapped using
getBootstrapSamples().
For this example, only 10 samples are bootstrapped for each model fit.
Show code
##Bootstrap quantiles
set.seed(7015) # re-sets seed only for this example; remove in your analysis script
bootstrap_quantiles <- getBootstrapQuantiles(
model_fits = model_fits,
quantiles = c(0.025, 0.5, 0.975),
doses = dose_levels,
n_samples = 10)Assessment of the Minimally Efficacious Dose
The Minimally Efficacious Dose (MED) per model shape can be assessed
with the function getMED(). The effect needs to be
specified on the probability scale.
Additional Note
Testing, modeling, and MED assessment can also be combined via
performBayesianMCPMod():
Show code
BMCPMod_result <- performBayesianMCPMod(
posterior_list = PostLogit,
contr = contr_mat_prior,
crit_prob_adj = crit_pval,
simple = TRUE,
delta = 0.16,
probability_scale = TRUE
)